Scroll to:
Comparative analysis of the genomic diversity of SARS-COV-2 circulating in the territories of the Rostov region and the republic of Crimea in the period from March to June 2021
https://doi.org/10.21886/2219-8075-2021-12-3-62-71
Abstract
Objective: To compare the genomic diversity of SARS-CoV-2 strains that were spread in the Rostov region and the Republic of Crimea in the period from March to June 2021.
Materials and Methods: A total of 194 samples were sequenced and 186 genovariants were recognized among them using the Pangolin program. Multiple alignment was performed and dendrograms were constructed for the samples belonging to the alpha and delta genovariants.
Results: Changes of the predominant genovariants were revealed for territories of the Rostov Region and the Republic of Crimea from March to June. An increasing percent of the alpha variant was observed in both regions during the spring followed by the rapid emergence of the delta variant, which became predominant in June.
Conclusion: It was shown that alpha variant samples from the Rostov region share a pool of common mutations, while in contrast, alpha variant samples from the Republic of Crimea are closer to those sampled from Moscow and Moscow region. At the end of May, the delta gene variant begins to be revealed, which is rapidly replacing other lines in all the territories considered in this study.
For citations:
Noskov A.K., Podoynitsina O.A., Vodopianov A.S., Pisanov R.V., Kovalev E.V., Penkovskaya N.A., Slis S.S., Yanovich E.G., Kuznetsova D.A., Vodopianov S.O., Chemisova O.S. Comparative analysis of the genomic diversity of SARS-COV-2 circulating in the territories of the Rostov region and the republic of Crimea in the period from March to June 2021. Medical Herald of the South of Russia. 2021;12(3):62-71. (In Russ.) https://doi.org/10.21886/2219-8075-2021-12-3-62-71
Introduction
More than a year and a half has passed since the start of the SARS-CoV-2 pandemic. By June 2021, there were more than 173 million confirmed cases of the novel coronavirus infection in the world, of which more than 3.7 million were fatal1. The widespread development of genome-wide sequencing methods and the creation of public databases containing data on the nucleotide sequences of SARS-CoV-2 viruses make it possible to study the features of the circulation of individual clones of the pathogen in different territories [1][2][3].
The genome length of SARS-CoV-2 is about 30 thousand base pairs [4]. It contains 14 open reading frames encoding both non-structural proteins (NSPs), responsible for the processes of viral replication and assembly, and structural proteins, including spike protein (S), envelope protein (E), membrane/matrix (M)б and nucleocapsid (N), as well as auxiliary proteins. The first reading frame contains approximately 65% of the viral genome and is translated into either the pp1a polypeptide (nsp 1-11) or the pp1ab polypeptide (nsp 1-16). Other frames encode structural and accessory proteins. Protein S is a transmembrane protein that facilitates binding of the viral envelope to angiotensin converting enzyme 2 (ACE2) receptors expressed on the surface of host cells. From the point of view of the pathogenesis of the disease, the most significant component of the coronavirus genome is the receptor-binding domain located in the virus spike protein gene and responsible for binding to the human cell receptor ACE2, thus participating in the penetration of the virus into cells. Protein N is involved in RNA replication, virion formation, and immune evasion. Protein M is one of the most conserved in the structure of virion proteins. It facilitates the assembly of viral particles. Protein E is the smallest component in the structure of SARS-CoV-2, which facilitates the production, maturation, and release of virions [5][6][7].
The WHO currently identifies four lines of SARS-CoV-2 that are classified as Variants of Concern (VOCs). In accordance with the PANGO nomenclature, these include lines B.1.1.7 (British genovariant), B.1.351 (South African genovariant), P.1 (Brazilian genovariant), and B.1.617.2 (Indian genovariant). They correspond to the WHO designations such as alpha, beta, gamma, and delta [8]. The alpha line, first identified in September 2020, by April 2021 held the leading position among all identified variants. At the same time, by June it was replaced by the SARS-CoV-2 strains belonging to the Pango B.1.617.2 line, which is of Indian origin.
Variants alpha, beta, gamma, and delta are characterized by certain sets of mutations in the receptor-binding domain of the spike protein, which affect the binding of the viral envelope to the ACE2 receptors [5][6] and can be associated with evasion of the immune response of the host, which can provide such strains have an evolutionary advantage and promote their rapid spread. The emergence of new lines of SARS-CoV-2 with possibly modified properties underscores the importance of monitoring existing variants.
The purpose of the study is a comparative analysis of the genomic diversity of variants circulating in the territories of the Rostov Region and the Republic of Crimea from March to June 2021.
Tasks:
- Determine the main lines allocated in the territories of the Rostov Region and the Republic of Crimea in the period from March to June 2021.
- Determine the presence/absence of "British", "South African", "Brazilian", and "Indian" genetic variants in these territories.
- Conduct a comparative analysis of the data obtained with the data of the Gisaid database in Moscow, the Moscow Region, and Europe.
- Compare the nucleotide sequences of genovariants belonging to line B.1.1.7 and detected in the Rostov Region, as well as in the Republic of Crimea, with the samples identified in Moscow and the Moscow Region.
- Compare the nucleotide sequences of genovariants belonging to line B.1.617.2 and detected in the territory of the Rostov Region, as well as in the territory of the Republic of Crimea, with the samples identified in the territory of Moscow and the Moscow Region.
Materials and Methods
For the analysis, 203 samples of biological material were used, the presence of the SARS-CoV-2 virus in which was confirmed by polymerase chain reaction (PCR). The AmpliSens® Cov-Bat-FL reagent kit was used to confirm the presence of the virus. Isolation of RNA was performed using the AmpliSens® RIBO-prep reagent kit. The preparation of cDNA was carried out using the AmpliSens® Reverta kit; primers proposed by the Artic consortium were used to generate specific amplicons. The primers were synthesized at NPF Syntol LLC (Moscow).
The amplification protocol proposed by the Artic consortium is based on fragmentary amplification of the SARS-CoV-2 virus genome using 98 primer pairs combined into two pools. However, according to reports of a number of authors [9][10], one of the problems in this case is the formation of primer dimers, which reduce the amplification efficiency. In this regard, the authors of this paper took the path of increasing the number of pools, dividing each of the pools into two more equal groups. Visual assessment of the results of electrophoresis showed more pronounced target bands (about 400 bp) and less pronounced primer dimers (less than 100 bp) when using four pools compared with two primer pools, which contributed to a better production of the target amplification product and gave subsequently more complete genome coverage. After amplification with subsequent sequencing, the research team managed to obtain 194 virus genomes, of which 193 were covered with more than 90% coverage. In this case, a coverage of 100% was obtained for 64 samples.
The PCR was carried out in a volume of 25 μl in polystyrene microcentrifuge tubes on a programmable multichannel thermal cycler Tertsik (DNA technology, Moscow).
The incubation mixture for PCR contained 20 mM Tris-HCl, pH 8.6; 7 mM MgCl2, 10 mM (NH4)2SO4, 0.5mM EDTA, 100 μg/ml BSA, 250 μM each of deoxynucleoside triphosphates, 15 nM of the corresponding primer, 2 units of Taq polymerase.
The amplicons were detected in a 1% agarose gel 10 cm long at a voltage of 220 V.
Sequencing was performed on a MiSeq Illumina instrument using a Nextera DNA Flex kit and a 500 cycle cartridge2. The assembly of genomes was carried out by alignment to the reference sequence of the hCoV-19/Wuhan/WIV04/20193 strain using the minimap2 [11], SAMtools [12], and iVar [13] programs.
Genetic lines were determined using the Pangolin software4.
Multiple alignment was performed for the entire genome and was implemented using the MAFFT program [14]. Phylogenetic trees were constructed using FastTree [15]. Visualization was performed using iTOL5. For comparative analysis, the data of whole genome sequencing was obtained from the GISAID database [16].
Results
The quantitative characteristics of the results of determining the SARS-CoV-2 genovariants in the studied samples (using the Pangolin program) from the territories of the Rostov Region (RR) and the Republic of Crimea (RC) are shown in Table 1.
Table 1
The frequency of detection of gene variants in the territories of the Rostov Region and the Republic of Crimea in the period from March to June 2021
|
Pango line |
Rostov Region |
Republic of Crimea |
Total in abs. numbers (RR + RC) |
||
|
Abs. number |
% |
Abs. number |
% |
||
|
B.1.1 |
38 |
36.5 |
55 |
61.1 |
93 |
|
B 1.1.7 |
25 |
24.0 |
11 |
12.2 |
36 |
|
B.1.617.2 |
10 |
9.6 |
10 |
11.1 |
20 |
|
B.1.1.130 |
8 |
7.7 |
0 |
0.0 |
8 |
|
B.1.202 |
0 |
0.0 |
3 |
3.3 |
3 |
|
B.1.95 |
2 |
1.9 |
1 |
1.1 |
3 |
|
B.1.1.4 |
3 |
2.9 |
0 |
0.0 |
3 |
|
B.1.1.220 |
2 |
1.9 |
0 |
0.0 |
2 |
|
B.1 |
0 |
0.0 |
1 |
1.1 |
1 |
|
B.1.1.317 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.351 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.292 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.1.151 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.1.315 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.1.216 |
0 |
0.0 |
1 |
1.1 |
1 |
|
B.1.247 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.1.194 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.1.102 |
0 |
0.0 |
1 |
1.1 |
1 |
|
B.1.177.8 |
0 |
0.0 |
1 |
1.1 |
1 |
|
B.1.1.261 |
1 |
0.9 |
0 |
0.0 |
1 |
|
B.1.1.141 |
0 |
0.0 |
1 |
1.1 |
1 |
|
None |
8 |
7.7 |
5 |
4.8 |
13 |
|
|
104 |
100 |
90 |
100 |
194 |
The dynamic results of similar content for the same time period using data for Moscow and for European countries for subsequent comparative analysis are presented in Figure 1.
Based on the results of the analysis of genomes using the Pangolin program, it was found that the largest number of SARS-CoV-2 identified in the territories of the Rostov Region and the Republic of Crimea for the entire research period (March – June 2021) belonged to the line "B.1.1" (Table 1).
According to the obtained data, this group of genovariants was most widely represented in March (57.7% in the Rostov Region and 81.8% in the Republic of Crimea) and April (29.6% and 71.4%, respectively) in 2021 (Table 2). In the following months, line B.1.1. was detected with a lower frequency, which was a consequence of an increase in the number of isolates of other lines (Figure 1).
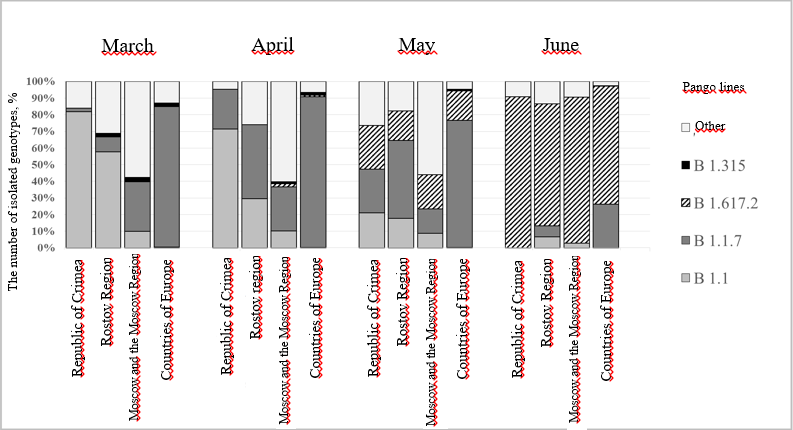
Figure 1. Pango lines (in %) allocated in the territories of the Rostov Region and the Republic of Crimea from March to June 2021
The displacement of B.1.1. occurred, in particular, due to a rapid increase in the genovariant alpha in the Russian Federation, as well as to the appearance of isolates of B.1.617.2. The graph (Figure 1) shows that in the territories of the Rostov Region and the Republic of Crimea, alpha line isolates appear in March (the frequency of detection is 8.9% and 2.3%, respectively). At the same time, already in May in the Rostov Region, among all the identified lines, line B.1.1.7 reaches 47.1%, and in Crimea, this value is 26.3% (Table 2). During the same period, isolates of the delta line were recorded for the first time in Moscow and the Moscow Region, the number of which continues to grow from April to the present (June 2021). In the Rostov Region and the Republic of Crimea, the "British" gene variant was first discovered in March, and by May the frequency of detection of representatives of this lineage reached its maximum values in both territories (47.1% and 26.3%, respectively), and in June their number was rapidly decreased to 6.7% for the Rostov Region and 0% for the Republic of Crimea. Isolates of line B.1.617.2 in the Rostov Region and the Republic of Crimea first appeared in May, and by June their number increased rapidly (73% in the Rostov Region and 91% in the Republic of Crimea ) (Table 2) and until now (June 2021) this line occupies a dominant position (Figure 1).
Table 2
The frequency of occurrence of VOC variants in the Rostov Region and the Republic of Crimea territories, as well as Moscow, the Moscow Region, and Europe
|
month
|
Genotypes that raise suspicion |
|||||||||||
|
B.1.1 |
B.1.1.7 |
B.1.617.2 |
||||||||||
|
RR* |
RC** |
Msc*** |
Eur**** |
RR |
RC |
Msc |
Eur |
RR |
RC |
Msc |
Eur |
|
|
March |
57.7% |
81.8% |
9.8% |
0.5% |
8.9% |
2.3% |
29% |
84.4% |
0.0% |
0.0% |
0.0% |
0.0% |
|
April |
29.6% |
71.4% |
10.2% |
0.2% |
44.4% |
23.8% |
26.5% |
90.7% |
0.0% |
0.0% |
1.9% |
1.0% |
|
May |
17.6% |
21.1% |
8.8% |
0.2% |
47.1% |
26.3% |
14.7% |
76.5% |
17.6% |
26% |
20.5% |
17.7% |
|
June |
6.7% |
0.0% |
2.8% |
0.1% |
6.7% |
0% |
0.0% |
26.2% |
73.3% |
90.9% |
87.9% |
70.9% |
** Republic of Crimea
*** Moscow and the Moscow Region
**** European countries
An increase in the share of line B.1.1.7 by April 2021 with its subsequent decrease in May also occurred in Europe. In January, in the Gisaid database (data from the Gisaid database, which are not included in Table 2) among all genovariants present in Europe, line B.1.1.7 occupies 52.8%. By April, this figure had grown to 90.7%, and in May it dropped to 76.5%. At the same time, the "Indian" variant in March was found in European countries in the amount of 0.01%, and by June its share was 70.9% (Table 2).
To compare the sequences of alpha and delta variants circulating in the territories of Moscow and the Moscow Region, as well as the Republic of Crimea and the Rostov Region, in the period from May to June 2021, dendrograms were constructed separately for each of the genetic lines (Figures 2, 3).
Discussion
When analyzing the results obtained, it was revealed that displacement B.1.1. occurred, in particular, due to an increase in the alpha genovariant on the territory of the Russian Federation, as well as the appearance and subsequent rapid growth of the number of representatives of the line B.1.617.2 (Figure 1). In the territories of the Rostov Region and the Republic of Crimea, alpha line isolates appeared in March (the frequency of detection is 8.9% and 2.3%, respectively). In total, 36 isolates belonging to variant B.1.1.7 and 20 – belonging to the genovariant delta were found in the above territories (Table 1).
When comparing the results obtained with the data of the Gisaid database, according to which line B.1.1.7 was detected in Moscow and the Moscow Region in January with a frequency of 16.3%, and then the proportion of detecting these isolates was growing, reaching 29.0% by March (database data), it can be seen that the trend towards an increase in the frequency of detection of isolates of the alpha variant is also characteristic for the territories of the Rostov Region and the Republic of Crimea, but with some lag. Apparently, this is due to the fact that, being an economic and transport center, the capital has a high intensity of migration processes, both international and domestic, contributing to the spread of SARS-CoV-2 and an increase in the incidence of COVID-19 in the peripheral regions of the country. According to the Gisaid database, the maximum number of "British" gene variants in Moscow and the Moscow Region was found in March, and in April and further, a gradual decrease in the representatives of this line was observed (Table 2).
Thus, in the countries of Europe, in the territories of Moscow and the Moscow Region, the Rostov Region, and the Republic of Crimea, from March to June 2021, there was a change in the dominant genetic variants. At the beginning of spring, the frequency of detection of isolates belonging to the alpha line increased, and then the delta genovariant rapidly displaced other lines in all territories considered in this study, which, in the authors’ opinion, is associated with its higher contagiousness.
Comparing the genetic sequences of "British" isolates identified in the Rostov Region, the Republic of Crimea, as well as Moscow and the Moscow Region, it was found that the vast majority of isolates of line B.1.1.7 identified in the Rostov Region during the period under consideration were located on the dendrogram as a separate cluster. Only one isolate (N 312 Bataysk on April 1, 2021) did not enter this cluster, making up one branch with isolates from the Moscow Region. The results obtained indicate the internal circulation of this etiological agent on the territory of the region against the background of the absence of cases of its import from other subjects of the Russian Federation and from abroad or the presence of isolated cases of import from other territories without further spread among the population of the Rostov Region (Figure 2).

Figure 2. Dendrogram. Comparison of isolates of the B.1.1.7 line detected in the Rostov Region (orange color), the Republic of Crimea (yellow color), in Moscow and the Moscow Region.
The maximum number of SARS-CoV-2 belonging to the B.1.1.7 lineage for the Rostov Region and the Republic of Crimea fell on May, and then the number of these isolates decreased rapidly. It can be assumed that in the spring, most of the passenger traffic is made up of people traveling on business trips, since the holiday season has not yet arrived. At the same time, the alpha line isolates obtained on the territory of Crimea are located in separate small clusters, which indicates both the presence in Crimea of isolates circulating for some time in this territory, and about multiple deliveries to this territory from the capital region.
Isolates of line B.1.617.2 in the Rostov Region and the Republic of Crimea first recorded in May, and by June their number increased rapidly (73% in the Rostov Region and 91% in the Republic of Crimea). Isolates that appeared in Crimea in May are located on the dendrogram in one cluster, which may be due to the introduction of the pathogen followed by local spread (Figure 3). In June, there is a rapid increase in the number of "Indian" genetic variants, which coincides with the beginning of the holiday season. The June isolates of the delta line identified in Crimea, as well as some May isolates of this line, are located on the dendrogram or separately in different clusters, thus forming common branches with both Moscow and isolates identified in the Rostov Region, or locally, which testifies to constantly occurring drifts. With the onset of the holiday season, the spread of the virus has increased among the susceptible population. It is likely that it will continue to grow until the end of the tourist season.

Figure 3. Dendrogram. Comparison of samples of the B. 1.617.2 line detected in the Rostov Region (orange color), the Republic of Crimea (yellow color), as well as in Moscow and the Moscow Region.
Conclusions
- In the period from March to June 2021, in the territories of the Rostov Region and the Republic of Crimea, there was a gradual change in the dominant SARS-CoV-2 lines with a predominance of B.1.1 in March, an increase in the proportion of alpha line isolates in April and May, and then the appearance of and a rapid increase in delta isolates by June in both territories. The only isolate of line B.1.315 was found on the territory of the Rostov Region, and representatives of line P.1 were not found.
- Comparing the obtained data with the data of the Gisaid database, it can be concluded that the change in the dominant genovariants took place both on the territory of Moscow and the Moscow Region, and on the territory of Europe, but at an earlier date. Apparently, the main gateway for the import of new lines to the territory of Russia is the capital, as the main logistics center of the country. And the spread of isolates inland is generally carried out with a certain delay, which makes it possible to predict the situation in the regions.
- The “British” variants identified in the Rostov Region apparently circulated in this area for some time and accumulated a pool of common mutations, while the isolates of this line, identified in Crimea, showed genome similarity with various isolates identified in Moscow and the Moscow Region, which indicates a series of independent deliveries of these viruses to the territory of the republic.
- Most of the isolates of the "Indian" variant identified in late May and early June in Crimea differ from those in Moscow and probably circulated for some time in Crimea, while B.1.617.2, isolated in June, as well as isolates found in the Rostov Region, show more similarities with the Moscow ones.
- The spread of line B.1.617.2 observed in the summer period has already led to an increase in the incidence. If the population does not comply with personal and public safety measures, as well as in case of untimely vaccination, it is likely that the number of patients may still increase, reaching a peak in September-October, when the vacation season ends and the bulk of the population will return to organized groups at the place of study and work.
1. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---1-june-2021
2. https://support.illumina.com/sequencing/sequencing_kits/nextera-dna-flex-kit/documentation.html
3. GenBank Accession Number MN908947.3
4. https://github.com/cov-lineages/pangolin
5. https://itol.embl.de
References
1. Kaptelova V.V., Speranskaya A.S., Samoilov A.E., Valdochina A.V., Bulanenko V.P. et al. Mutations in the genomes of Sars-CoV-2 from clinical samples obtained in late March – early April from patients in Moscow. Russian national scientifi c and practical conference with international participation “Molecular Diagnostics and Biosafety – 2020”; October 6–8, 2020; Moscow. (In Russ) doi: 10.36233/978-5-9900432-9-9-147
2. Kosyreva A.N., Bakshtanovskaya I.V., Stepanova T.F., Letyshev A.N., Stepanova K.B. Results of detection Sars-CoV-2 and other pathogens of community-acquired pneumonia by PCR in the Tyumen oblast in April – July 2020. Russian national scientifi c and practical conference with international participation “Molecular Diagnostics and Biosafety – 2020”; October 6–8, 2020; Moscow. (In Russ) doi:10.36233/978-5-9900432-9-9-153
3. Nagy Á, Pongor S, Győrff y B. Diff erent mutations in SARS-CoV-2 associate with severe and mild outcome. Int J Antimicrob Agents. 2021;57(2):106272. DOI: 10.1016/j.ijantimicag.2020.106272
4. Devika S., Soojin V. Yi. On the origin and evolution of SARSCoV- 2. Exp Mol Med. 2021;53(4):7–547. DOI: 10.1038/s12276-021-00604-z
5. Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J Microbiol Immunol Infect. 2021;54(2):159–163. DOI: 10.1016/j.jmii.2020.03.022
6. Wong N.A., Saier M.H. The SARS-Coronavirus Infection Cycle: A Survey of Viral Membrane Proteins, Their Functional Interactions and Pathogenesis settings. Int. J. Mol. Sci. 2021;22(3):1308. DOI: 10.3390/ijms22031308
7. Wang M., Zhao R., Gao L., Xue-Fei Gao, Wang D. et al. SARSCoV-2: Structure, Biology, and Structure-Based Therapeutics Development Front Cell. Infect Microbiol. 2020;10:587269. DOI: 10.3389/fcimb.2020.587269
8. Kannan S., Shaik Syed Ali P., Sheeza A. Evolving biothreat of variant SARS-CoV-2 – molecular properties, virulence and epidemiology. Eur Rev Med Pharmacol Sci. 2021;25(12):4405-4412. DOI: 10.26355/eurrev_202106_26151.
9. Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv [Preprint]. 2020:2020.09.04.283077. DOI: 10.1101/2020.09.04.283077
10. Itokawa K, Sekizuka T, Hashino M, Tanaka R., Kuroda M. Disentangling primer interaction simproves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS ONE. 2020;15(9):0239403. DOI: 10.1371/journal.pone.0239403
11. Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094-3100. DOI:10.1093/bioinformatics/bty191
12. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J., Homer N. et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25(16):2078-9. DOI: 10.1093/bioinformatics/btp352
13. Grubaugh N.D., Gangavarapu K., Quick J, Matteson N.L., De Jesus J.G. et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;8 DOI: 10.1186/ s13059-018-1618-7
14. Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. // Nucleic Acids Res. – 2002. – V.30. – P.3059 – 3066. DOI: 10.1093/nar/gkf436 14. Katoh K., Misawa K., Kuma K., Miyata T.) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059-3066. DOI: 10.1093/nar/gkf436
15. Price M.N., Dehal P.S., Arkin A.P. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE. 2010;5(3):e9490. DOI: 10.1371/journal.pone.0009490
16. Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenges. 2017;1:33-46. DOI: 10.1002/gch2.1018
About the Authors
A. K. NoskovРоссия
Alexey K. Noskov, Dr. Sci. (Med.), Director
Rostov-on-Don
O. A. Podoynitsina
Россия
Oksana A. Podoynitsina, Dr. Sci. (Biol.) researcher at the Laboratory of Microbiology of Cholera
Rostov-on-Don
A. S. Vodopianov
Россия
Aleksej S. Vodopianov, Dr. Sci. (Med.), senior researcher at the Laboratory of the diagnosis particularly dangerous Infections
Rostov-on-Don
R. V. Pisanov
Россия
Ruslan V. Pisanov, Dr. Sci. (Biol.) the acting head Laboratory of the diagnosis particularly dangerous Infections
Rostov-on-Don
E. V. Kovalev
Россия
Evgenij V. Kovalev, the Chief of Surveillance
Rostov-on-Don
N. A. Penkovskaya
Россия
Natalja A. Pen’kovskaja, the head of the Interregional
Simferopol
S. S. Slis
Россия
Sergej S. Slis’, leading researcher
Rostov-on-Don
E. G. Yanovich
Россия
Evgeniya G. Yanovich, Dr. Sci. (Med.), junior research associate, laboratory of epidemiology of particularly dangerous infections
Rostov-on-Don
D. A. Kuznetsova
Россия
Darja A. Kuznetsova, researcher at the Laboratory of Plague Microbiology
Rostov-on-Don
S. O. Vodopianov
Россия
Sergey O. Vodopianov, Dr. Sci. (Med.) acting head of the Laboratory of Biochemistry of microbes
Rostov-on-Don
O. S. Chemisova
Россия
Olga S. Chemisova, Dr. Sci. (Biol.) the acting head of the Museum of Living Cultures
Rostov-on-Don
Review
For citations:
Noskov A.K., Podoynitsina O.A., Vodopianov A.S., Pisanov R.V., Kovalev E.V., Penkovskaya N.A., Slis S.S., Yanovich E.G., Kuznetsova D.A., Vodopianov S.O., Chemisova O.S. Comparative analysis of the genomic diversity of SARS-COV-2 circulating in the territories of the Rostov region and the republic of Crimea in the period from March to June 2021. Medical Herald of the South of Russia. 2021;12(3):62-71. (In Russ.) https://doi.org/10.21886/2219-8075-2021-12-3-62-71
JATS XML







































