Scroll to:
Human coronaviruses that can cause emergencies
https://doi.org/10.21886/2219-8075-2021-12-1-14-23
Abstract
The first coronavirus NСoV-B814 was isolated from humans in 1965 and did not survive to the present time. For a long time, it was believed that coronaviruses were not pathogenic to humans. They were not included in the list of particularly dangerous infections and represented a serious problem exclusively in veterinary medicine. But in 2002, after the SARS outbreak, scientists’ opinions changed. A new subtype of the coronavirus called SARS-CoV penetrated the human population. In 2012, it was possible to discover natural foci of Middle East Respiratory Syndrome. The epidemic of a new coronavirus infection that emerged in late 2019 and early 2020 attracted the attention of scientists around the world. The priority was a detailed and close study of all the varieties of this virus. This review describes seven types of coronaviruses that can cause emergencies in populations around the world.
Keywords
For citations:
Kononenko A.A., Noskov A.K., Vodyanitskaya S.Yu., Podoynitsyna O.A. Human coronaviruses that can cause emergencies. Medical Herald of the South of Russia. 2021;12(1):14-23. https://doi.org/10.21886/2219-8075-2021-12-1-14-23
In the 1930s, after numerous studies, coronaviruses were officially acknowledged pathogenic for animals. Thirty years after, the strains were identified that caused respiratory diseases in humans [1].
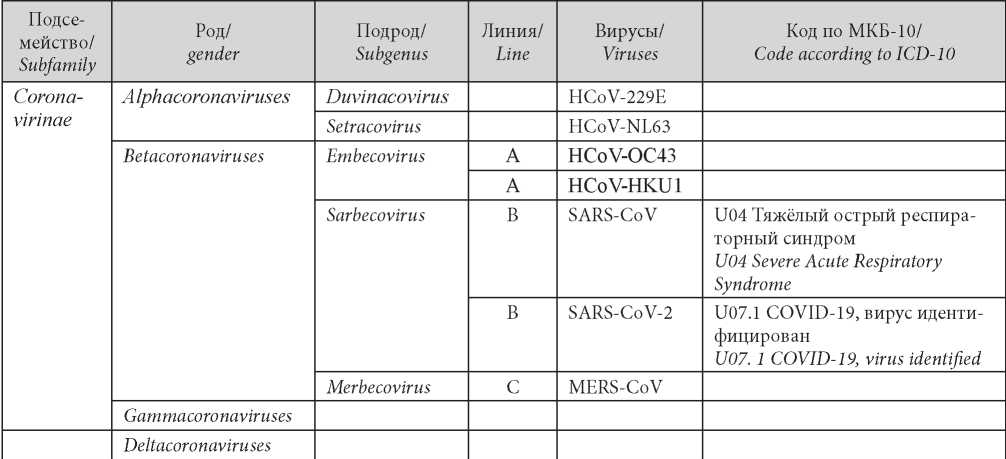
Presently, coronaviruses form the largest group of the order Nidovirales that includes such families as Coronaviridae, Arteriviridae, Roniviridae, and Mesoviridae. The Coronaviridae family consists of two subfamilies Coronavirinae and Torovirinae. Coronaviruses are divided into 4 genera: Alphacoronaviruses, Betacoronaviruses, Gammacoronaviruses, and Deltacoronaviruses. Initially, within the genus Betacoronavirus, the viruses were divided into lines A, B, C, and D that were later renamed Embecovirus (previous line A), Sarbecovirus (previous line B), Merbecovirus (previous line C), and Nobecovirus (previous line D) [2]. These 4 lines are classified into subgenera of Betacoronaviruses.
Among numerous representatives of the family, seven coronaviruses can cause diseases in humans (Table 1). Four viruses (HCoV-229E, HCoV-NL63, HCoVOC43, and HCoV-KHU1) are etiologic agents of acute respiratory viral infections (ARVIs) of light or moderate severity. Two viruses are capable of causing lethal diseases: severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV) and Middle East respiratory syndrome-related coronavirus (MERS-CoV). The 7th virus (SARS-CoV-2) is a novel coronavirus. The first case of infection was registered in China at the end of 2019. At the beginning of 2020, SARS-CoV-2 spread all over the world [3][4].
Table 1 / Таблица
Classification of Coronavirinae viruses
Классификация вирусов Coronavirinae
Human coronaviruses HCoV-229E and HCoV-OC43
In 1965, Tyrrell and Bynoe were the first to cultivate the virus that was isolated from the respiratory tract of a boy with regular cold by means of passage in the embryonal cultures of the organs of the human trachea [5][6]. The first official scientific publication dedicated to coronaviruses is dated November 16,1968 in the journal Nature. It was proposed in the article to unite these viruses into a group of “coronaviruses” because of a typical morphology of virions 229E and OC43 with a precisely expressed corona-like enclosure (20 nm) of roundish pleiomorphic particles (120–160 nm) [7].
By a degree of pathogenicity, Coronaviridae were classified as
- Group IV of pathogenicity that included HCoV229E and numerous animal viruses;
- Group III of pathogenicity that included viruses HCoV-OC43 and closely related animal viruses [6].
According to the official data obtained from different studies on volunteers included in the “healthy group”, it was established that viruses HCoV-229E and HCoVOC43 caused a regular cold [8][9]. Since then, HCoVs were considered relatively pathogenic respiratory viruses.
It was proved experimentally that HCoV-229E and HCoV-OC43 were characterized by droplet transmission. Volunteers infected with this virus produced it within 5 days starting 48 hours after infecting, which corresponded to the appearance of the first symptoms of the disease [10].
The studies showed that infection HCoV-229E was performed by the inoculation of the mucous membranes of the respiratory tract. The exudation of plasma of the nasal mucosa and increased levels of interferons γ (IFNy) in the samples of nasopharyngeal lavage directly depended on the severity of symptoms [11].
The viral load of the respiratory tract reached its maximum within the first three days after the infection and sharply decreased one week later, which was associated with the appearance of acquired active immunity in humans [12].
Starting from 2000, representatives of the Coronaviridae family were revealed that caused severe respiratory diseases that could result in a lethal outcome.
Human coronavirus SARS-CoV
The first case of the disease caused by SARS-CoV was registered in Shanlan, Guangdong province, in the south of China in November 2002. The transmission of the infection primarily occurred in hospital conditions. On average, one infected patient infected 3–4 people that they contacted. Italian doctor Carlo Urbani, who was a member of “Doctors Without Borders”, was the first to identify the observed disease as novel. Carlo Urbani got infected during the treatment of a patient with SARS and died. The strain of the isolated virus was called after this doctor SARS-related human coronavirus Urbani (SARS). Due to Carlo Urbani and his colleagues’ activities, it was possible to stop the spread of SARS [4].
In 2003, it was shown that the infecting agent of the disease was an earlier unknown variant of coronavirus. It was named SARS–CoV. Its genome organization was similar to coronavirus. However, phylogenetic analysis and comparison of the sequences suggested that SARS–CoV was not related to any of the viruses described before. Its virions contained a plus-strand of polyadenylated RNA 16–30 kb long (coronaviruses have the largest genome that threefold exceeds the genome of other viruses) [13]. SARS-CoV was classified as Group III pathogenicity accepted in the Russian Federation.
Based on the conducted studies, a hypothesis was made in the primary penetration of the virus into the human population when they ate civet meat because coronavirus isolated from these animals was inactivated by the blood serum of patients with SARS-CoV [14][15].
Further, it was established that natural foci of SARS-CoV virus were associated with the habitats of Chiroptera, primarily, bats. Besides, SARS-CoV-like viruses were isolated from horseshoe bats Rhinolophus and species that belonged to other genera found in Asia (primarily, China), Africa, Australia, Europe, and the USA. It is impossible to exclude direct transmission of the infection to humans via the wastes of bats that can live in the attics of living facilities [3][16].
The severity of the disease varied from a light to severe form with a lethal outcome. In the majority of patients, the disease developed to a light or moderate degree with signs of regular ARVI, quick recovery, and specific immunity. However, some patients developed a severe form of pneumonia, associated with severe acute respiratory syndrome [17][18].
The spread of the disease was stopped in 2003 by highly effective global measures taken by the systems of healthcare worldwide. Presently, SARS does not circulate among the human population [19][20].
As it was noted above, coronaviruses are capable of evolving and adapting to new hosts. The outburst of atypical pneumonia returned coronaviruses to the center of researchers’ attention. As a result, two more representatives of the Coronaviridae family that could cause diseases in humans were isolated.
Human coronaviruses HCoV-NL63 and HCoVHKU1
In 2004, Dutch scientists in the Netherlands were the first to isolate a novel virus HCoV-NL63 (Human coronavirus NL63) from a seven-month-old child with bronchiolitis. This virus was classified as Group IV of pathogenicity [21][22][23].
In January 2005, researchers from the University of Hong Kong isolated a novel human coronavirus HKU1 (HCoV-HKU1 — Human coronavirus HKU1) from a 71-year-old patient with acute respiratory disease complicated with bilateral pneumonia [24]. Hong Kong University introduced the prefix HKU in the classification of coronaviruses with a number of the strain that is met in the names of numerous viruses. HCoV-HKU1 virus was classified as Group III of pathogenicity.
HCoV-NL63 and HCoV-HKU1 are one-strand positive RNA viruses. The majority of patients had such symptoms as rhinorrhea, fever, cough, and hissing respiration. The disease manifestations included bronchitis and pneumonia [25][26].
The first cases of infection of people with HCoVNL63 were registered among infants with a severe infection of the lower respiratory tract in the inpatient conditions, while the first cases of HCoV-HKU1 infection were registered among senior patients with the main comorbid diseases of the respiratory and cardiovascular systems. Presently, HCoV-HKU1 is also registered in children with acute respiratory diseases and infections of the upper or lower respiratory tracts [21][26].
As a rule, diseases caused by HCoV-HKU1 and HCoV-NL63 viruses are not life-threatening, especially, for healthy people. It was suggested that HCoV-NL63 and HCoV-HKU1, as well as HCoV-229E and HCoVOC43, are capable to provoke diseases with expressed clinical symptoms only in children, senior people, and those who have immune disorders [27].
From 2006 to 2012, coronaviruses were actively studied. In June 2012, a new representative of this family was isolated, which was called MERS-CoV.
Human coronavirus MERS-CoV
In June 2012, in Jidda (Saudi Arabia), a virologist Ali Mohamed Zaki isolated a novel coronavirus from the sputum of a patient who died because of severe viral pneumonia complicated by an acute kidney injury. By many signs, the virus was close to SARS-CoV. Thus, the novel infection was called Middle East respiratory syndrome.
Initially, the virus MERS-CoV was planned to be called a “virus of acute respiratory syndrome with kidney injury” but it was soon revealed that kidney injury was not a leading pathology, so for some time, the term “novel coronavirus” (HCoV-EMC/2012) was used. In May 2013, the virus was called MERS-CoV by the International Committee on Viral Taxonomy [28].
Even though the transmission of a virus between people was not intensive, MERS-CoV caused two major outbursts in Saudi Arabia (2012) and South Korea (in 2015), wherein the general number of confirmed cases exceeded two thousand at the level of lethality 35% [29]. In senior patients, especially those who had comorbid pathology, the infection caused by MERS-CoV had a sore severe development and often had a lethal outcome [28].
The virus MERS-CoV is a linear non-segmented one-strand positive RNA. By the degree of pathogenicity, the virus was classified as Group II by the classification accepted in the Russian Federation.
It was suggested that bats could be not the only reservoir of MERS-CoV. Some data confirmed the possibility of carriership in Erinaceus europaeus. Besides, it is possible that camels can host MERS-CoV [30].
It was established that MERS-CoV strains isolated from camels were completely identical to the strains isolated from humans. Besides, antibodies specific to MERS-CoV were often found in the samples of serum obtained from camels from the Middle East, Africa, and Asia collected in 1983. This, it is suggested that viruses circulate in the blood of camels for more than 30 years.
Numerous studies officially confirmed that the natural reservoir for MERS-CoV is a bat (Chiroptera). The virus isolated from one of the patients was identical in the molecular-genetic aspect to the virus isolated from pouched bats (Taphozous perforatus) from the family of shelf-tailed bats (Emballonuridae) [31]. Bats expressed virus with saliva, urine, and feces, that could become the source of infection for humans and other animals. Pouched bats were met not only in the south of the Arabian Peninsula but also in the western part of the Hindustan peninsula. Thus, it cannot be excluded that the areas of natural foci of MERS-CoV were wider than it was believed. It was shown that MERS-CoV was capable to reproduce in the primary cellular structures obtained from bats in different taxonomic groups: short-tailed leaf-nosed bats (Phyllostomidae, Carollia) and rousettes (Pteropodidae, Rousettus) [32].
Researchers from Saudi Arabia conducted a serological investigation in the territory of Oman to reveal MERS-CoV in the populations of farm animals. It showed that 100% of dromedaries (Camelus dromedarius) had antibodies against subunit S1 of the spike protein MERS-CoV. Further, direct evidence was obtained of the circulation of MERS-CoV variants identical to the epidemic ones in the organism of camels and the possibility of infection of humans from these animals [33]. Wing-handed animals contaminated camels during their day’s rest in the barns for livestock. It was revealed that there were no specific antibodies in dromedaries to MERS-CoV in Africa, including Canary Islands [32]. Along with this, specific antibodies to MERS-CoV lacked in dromedaries in Australia [34], which indicated that these animals could not be the main host of MERS-CoV. Specific MERS antibodies were revealed in alpacas from Qatar (Vicugna pacos) [35]. Probably, all Tylopoda (Artiodactyla: Tylopoda) are sensitive to virus MERS-CoV and can be an intermediate link between the host and indicator of this virus if there is a natural reservoir –Chiroptera that contain the virus. This suggestion agrees with the lack of specific immunity in Bactrian camels (Camelus bactrianus) in non-endemic for MERS-CoV territories in Kazakhstan [36], Mongolia [37], and Northern China [38].
Even though the search for hosts of MERS-CoV among farm animals (bovine cattle, horses, goats, sheep) did not bring results, these studies significantly stimulated the study of coronaviruses in the populations of mammals apart from Chiroptera. There is evidence of transmission of MERS-CoV from humans to humans and the only source of infection is an infected human. However, the transmission of MERS-CoV from a human to a human occurs only after close contact with an infected person, for example, in medical institutions or when the secondary infection is added.
Presently, in Saudi Arabia, MERS-CoV is a serious threat to the human population because millions of religious people from 184 countries travel to Hadj or Umru, which can provoke new epidemics with a wider areal of spread. The last case of infection with MERSCoV was registered in Saudi Arabia in February 2020 in a senior man with several chronic diseases.
Coronavirus infection SARS-CoV-2
At the end of 2019, a novel lethal virus appeared in the People’s Republic of China (PRC). The virus caused pneumonia in Hankow, Hubei province, China. The quick spread of the virus started the epidemics in China, which was followed by a pandemic. In February 2020, the World Health Organization (WHO) called the infection “COVID-19” (COronaVIrus Disease 2019)1.
On February 11, 2020, the International Committee on the Taxonomy of Viruses called the novel coronavirus SARS-CoV-2. The genetic sequence of SARS-CoV-2 was similar to the sequence SARS-CoV at least by 79%.
The analysis of the genetic basis of S-protein Spike, conducted by the specialists of the leading scientific centers in the USA, Australia, and Great Britain, showed that because of the occurred mutations in the receptor-binding domain of spike-like proteins, SARSCoV-2 (unlike other coronaviruses) acquired a capability of binding to ACE2 receptor. It was suggested that for such an uptake, ACE2 also had to undergo certain genetic changes. This shows that the capability of SARSCoV-2 to infect humans is the result of natural selection [39][40][41].
The first cases of the disease were associated with the sea product market in Hankow that sells poultry and meat of exotic animals (bats, snakes, pangolins). However, the specialists from the tropical botanical garden in China Academy of Sciences concluded that it was not the primary focus of the disease spread because the first cases of infection with novel coronavirus were registered in November 2019 and were not connected [42]. Besides, an epidemiological association was established between the trips of patients to PRC from neighboring countries. The largest number of infected people was revealed in the southern-eastern part of China (84% of the total cases in PRC) [43].
SARS-CoV-2 is a one-strand RNA-containing virus. It is Group II by the classification of pathogenicity. SARS-CoV-2 is included in the list of dangerous diseases.
The genetic assay of the virus that caused the disease showed similarity with coronaviruses spread among horseshoe bats. However, it is still not known if the bats were the initial source of infection. Nowadays, the main pathway of infection is human to human.
Scientists quickly established that SARS-CoV-2 was highly contagious and it could survive up to 2 hours in the environment and from several hours to 2 days on the surfaces [44]. The incubation period after infection is up to 14 days. All age groups are susceptible to the virus. Senior patients with comorbid diseases are more prone to a severe form. The main sources of infection are the diseased, symptomless carriers, and patients in the incubation period. Until now, the main pathways of infection were droplet and contact. The vertical pathway between the mother and newborn was suspected when a newborn was diagnosed with the novel coronavirus infection 30 hours after the birth in a hospital in Hankow. Besides, it is suggested that infection gets to the organism via the conjunctiva because conjunctival epithelium can be infected with aerosol or other biological liquids containing virus [45].
At the beginning of the epidemic spread, virus properties had a stable character with the clonal identity of the virus SARS-CoV-2. Mass-scale screening studies allowed the researchers to reveal the initial evolutionary changes in the virus in different parts of the world. All the viruses mutate with time and SARS-CoV-2 is not an exception.
The sequencing showed that coronaviruses changed slower than the majority of other RNA viruses because of an enzyme that fixes potentially fatal errors of copying. The typical SARS-CoV-2 virus accumulates only two single-letter mutations per month in its genome. This rate is twice as lower than in flu and ¼ lower than in HIV. This study was conducted at the Basel University, Switzerland2.
The study of the University College of London showed that in two viruses SARS-CoV-2 collected from different parts of the world, only 10 RNA letters out of 29,903 differed from each other. This allows specialists to monitor evolutionary changes in the virus. Lots of mutations are not significant for the capability of the virus to spread or cause diseases among humans. However, several genetic changes are evolutionary beneficial for the virus and promote its spreading capacity. For example, line B.1.1.7 (“British strain”) is a phylogenetic cluster that quickly spreads in the south-east of England. Before it was isolated at the beginning of September 2020, this line accumulated 17 specific mutations, which indicated a significant evolution, probably, in the chronically infected host. On December 28, 2020, this variant was detected in nearly 28% of cases of SARSCoV-2 infection in England, and population-genetic models suggest that this line spreads 56% quicker than others. Line B.1.1.7 spread when SARS-CoV-2 was revealed all over the world and started to dominate in the existing population of the circulating variants. This indicates a natural selection of the virus with higher transmissibility at the population level. The measures taken by public healthcare services (protective masks, social distancing, and limit on social gathering) should remain effective but the fight with a more transmissive (“more contagious”) variant will require stricter measures worldwide. Eight mutations in line B.1.1.7 involve spike glycoprotein (S-protein). It is suggested that these mutations can influence the binding of ACE2 and virus replication. Another coronavirus strain, also with a mutation in the receptor-binding domain, quickly spreads in South Africa. The influence of the mutation on antigenicity is still understudied3.
Since SAR-CoV-2 already spread in all the continents and the number of infected people is still growing, the primary task of the systems of healthcare all over the world is specific measures of prevention. In December 2020, different medical institutions and pharmaceutical companies were working on more than 200 potential vaccines.
On August 11, 2020, in Russia, a vaccine “GamCovid-Vac (Sputnik V)” was invented at the Scientific and Research Center of Epidemiology and Microbiology named after Gamaleya.
Later, on October 13, 2020, Russia announced the registration of another vaccine “EpiVacCorona” developed by the State Scientific Center of Virology and Biotechnology “Vector” (Rospotrebnadzor).
Another perspective variant that entered the stage of clinical studies is the whole-virion vaccine (Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of Russian Academy of Sciences, Moscow).
According to the WHO, in 2021, clinical studies of 63 candidate vaccines are carried out all over the world. It is proved that people have a high susceptibility to coronavirus: all age groups are prone to this disease. Antigen diversity of viruses provides a high rate of reinfection.
The appearance of new representatives of the coronavirus family provides the rationale for their further study to choose promising preventive (antiepidemic) measures, improve specific prevention, and develop vaccines.
The current generation witnessed a pandemic that is extraordinary for modern life. Presently, specialists all over the planet are working on effective measures against COVID-19. Quarantine measures and sanctions are introduced for their violation. Humankind has yet to evaluate and comprehend the scale of the socialeconomic damage and consequences of human losses. Time will show how society will change after the COVID-19 pandemic.
1. Kenneth McIntosh Coronavirus disease 2019 (COVID-19): Epidemiology, virology, and prevention https://www.uptodate.com/contents/coronavirusdisease-2019-covid-19-epidemiology-virology-and-prevention
2. Electronic resource https://www.nature.com/articles/d41586-020-02544-6
3.Electronic resource https://jamanetwork.com/journals/jama/fullarticle/2775006
References
1. Shchelkanov M.Yu., Popova A.Yu., Dedkov V.G., Akimkin V.G., Maleyev V.V. History of investigation and current classification of coronaviruses (Nidovirales: Coronaviridae). Russian Journal of Infection and Immunity. 2020;10(2):221-246. (In Russ.) DOI: 10.15789/2220-7619-HOI-1412
2. Li X, Luk HKH, Lau SKP, Woo PCY. Human Coronaviruses: General Features. Reference Module in Biomedical Sciences. 2019:B978-0-12-801238-3.95704-0. DOI: 10.1016/B978-0-12-801238-3.95704-0.
3. L’vov D.K., Kolobuhina L.V., Deryabin P.G. Coronavirus infection. Severe acute respiratory syndrome. Infekcionnye bolezni: Novosti. Mneniya. Obuchenie. 2015;4(13:35-42) (In Russ.). eLIBRARY ID: 25197472
4. Lvov D.K., Alkhovsky S.V., Kolobukhina L.V., Burtseva E.I. Etiology of epidemic outbreaks COVID-19 in Wuhan, Hubei province, Chinese People Republic associated with 2019- nCoV (Nidovirales, Coronaviridae, Coronavirinae, Betacoronavirus, Subgenus Sarbecovirus): lessons of SARS-CoV outbreak. Problems of Virology. 2020;65(1):6-15. (In Russ.) DOI: 10.36233/0507-4088-2020-65-1-6-15
5. Shamsheva O.V. New coronavirus NCOV-19 (SARS-COV-2). Detskie infekcii. 2020;19(1):5-6 (In Russ.). eLIBRARY ID: 42706398
6. Tyrrell DA, Bynoe ML. Cultivation of a novel type of commoncold virus in organ cultures. Br Med J. 1965;1(5448):1467-70. DOI: 10.1136/bmj.1.5448.1467
7. Almeida J.D., Berry D.M., Cunningham C.H., Hamre D., Hofstad M.S., Mallucci L., McIntosh K., Tyrrell D.A.J. Virology: Coronaviruses. Nature. 1968;220(5168):650. DOI: 10.1038/220650b0
8. Bradburne AF, Somerset BA. Coronative antibody tires in sera of healthy adults and experimentally infected volunteers. J Hyg (Lond). 1972;70(2):235-44. DOI: 10.1017/s0022172400022294
9. Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121(1):190-3. DOI: 10.3181/00379727-121-30734.
10. Akerlund A, Greiff L, Andersson M, Bende M, Alkner U, Persson CG. Mucosal exudation of fibrinogen in coronavirusinduced common colds. Acta Otolaryngol. 1993;113(5):642-8. DOI: 10.3109/00016489309135878
11. Linden M, Greiff L, Andersson M, Svensson C, Akerlund A, et al. Nasal cytokines in common cold and allergic rhinitis. Clin Exp Allergy. 1995;25(2):166-72. DOI: 10.1111/j.1365-2222.1995.tb01022.x
12. Shchelkanov M.YU., Kolobuhina L.V., L’vov D.K. Human coronaviruses (Nidovirales, Coronaviridae): the increased level of the epidemic danger. Lechashchij vrach. 2013;10:49-54. (In Russ.). eLIBRARY ID: 22592720
13. Chuchalin A.G. Severe acute respiratory syndrome (SARS). Russkij medicinskij zhurnal. 2003;11(194);1197-1204. (In Russ.).
14. Generalov I.I. ed. Medicinskaya virusologiya: uchebnoe posobie. Vitebsk, VGMU, 2017. (In Russ.).
15. Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995-4008. DOI: 10.1128/JVI.06540-11
16. Balboni A, Battilani M, Prosperi S. The SARS-like coronaviruses: the role of bats and evolutionary relationships with SARS coronavirus. New Microbiol. 2012;35(1):1-16. PMID:22378548.
17. Andrejchik M.A., Kopcha V.S., Nychik N.A. Severe acute respiratory syndrome. Mezhdunarodnyj medicinskij zhurnal. 2003;3:98-102. (In Russ.).
18. Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101(16):6212-6. DOI: 10.1073/pnas.0400762101.
19. Lednicky JA, Waltzek TB, McGeehan E, Loeb JC, Hamilton SB, Luetke MC. Isolation and genetic characterization of human coronavirus NL63 in primary human renal proximal tubular epithelial cells obtained from a commercial supplier, and confirmation of its replication in two different types of human primary kidney cells. Virol J. 2013;10:213. DOI: 10.1186/1743-422X-10-213
20. van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, et al. Identification of a new human coronavirus. Nat Med. 2004;10(4):368-73. DOI: 10.1038/nm1024
21. Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79(2):884-95. DOI: 10.1128/JVI.79.2.884-895.2005
22. Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44(6):2063-71. DOI: 10.1128/JCM.02614-05
23. Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35(1):99-102. DOI: 10.1016/j.jcv.2005.09.008
24. Arden KE, Nissen MD, Sloots TP, Mackay IM. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005;75(3):455-62. DOI: 10.1002/jmv.20288
25. Chiu SS, Chan KH, Chu KW, Kwan SW, Guan Y, et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40(12):1721-9. DOI: 10.1086/430301
26. Vabret A, Dina J, Gouarin S, Petitjean J, Corbet S, Freymuth F. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin Infect Dis. 2006;42(5):634-9. DOI: 10.1086/500136
27. Bastien N, Anderson K, Hart L, Van Caeseele P, Brandt K, et al. Human coronavirus NL63 infection in Canada. J Infect Dis. 2005;191(4):503-6. DOI: 10.1086/426869
28. Shchelkanov M.YU., Anan’ev V.YU., Kuznecov V.V., SHumatov V.B. Middle East respiratory syndrome: when will the smouldering fire break out? Tihookeanskij medicinskij zhurnal. 2015;2:94-98 (In Russ.). eLIBRARY ID: 24275106
29. Müller MA, Raj VS, Muth D, Meyer B, Kallies S, et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio. 2012;3(6):e00515-12. DOI: 10.1128/mBio.00515-12
30. De Benedictis P, Marciano S, Scaravelli D, Priori P, Zecchin B, et al. Alpha and lineage C betaCoV infections in Italian bats. Virus Genes. 2014;48(2):366-71. DOI: 10.1007/s11262-013-1008-x
31. Gortazar C, Segalés J. Middle East respiratory syndrome (MERS) coronavirus: a new challenge for veterinarians? Vet Pathol. 2013;50(6):954-5. DOI: 10.1177/0300985813506391
32. Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370(26):2499-505. DOI: 10.1056/NEJMoa1401505
33. Corman VM, Jores J, Meyer B, Younan M, Liljander A, et al. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013. Emerg Infect Dis. 2014;20(8):1319-22. DOI: 10.3201/eid2008.140596
34. Crameri G, Durr PA, Barr J, Yu M, Graham K, et al. Absence of MERS-CoV antibodies in feral camels in Australia: Implications for the pathogen’s origin and spread. One Health. 2015;1:76-82. DOI: 10.1016/j.onehlt.2015.10.003
35. Reusken CB, Schilp C, Raj VS, De Bruin E, Kohl RH, et al. MERS-CoV Infection of Alpaca in a Region Where MERSCoV is Endemic. Emerg Infect Dis. 2016;22(6):1129-31. DOI: 10.3201/eid2206.152113
36. Miguel E, Perera RA, Baubekova A, Chevalier V, Faye B, et al. Absence of Middle East Respiratory Syndrome Coronavirus in Camelids, Kazakhstan, 2015. Emerg Infect Dis. 2016;22(3):555-7. DOI: 10.3201/eid2203.151284
37. Chan SM, Damdinjav B, Perera RA, Chu DK, Khishgee B, et al. Absence of MERS-Coronavirus in Bactrian Camels, Southern Mongolia, November 2014. Emerg Infect Dis. 2015;21(7):1269-71. DOI: 10.3201/eid2107.150178
38. Liu R, Wen Z, Wang J, Ge J, Chen H, Bu Z. Absence of Middle East respiratory syndrome coronavirus in Bactrian camels in the West Inner Mongolia Autonomous Region of China: surveillance study results from July 2015. Emerg Microbes Infect. 2015;4(12):e73. DOI: 10.1038/emi.2015.73
39. Belotserkovskaia Yu.G., Romanovskikh A.G., Smirnov I.P. COVID-19: a respiratory infection caused by new coronavirus: new data on epidemiology, clinical course, and patients management. Consilium Medicum. 2020;22(3):12–20. (In Russ.). DOI: 10.26442/20751753.2020.3.200092
40. Xu X, Chen P, Wang J, Feng J, Zhou H, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457-460. DOI: 10.1007/s11427-020-1637-5
41. Jaimes JA, Millet JK, Stout AE, André NM, Whittaker GR. A Tale of Two Viruses: The Distinct Spike Glycoproteins of Feline Coronaviruses. Viruses. 2020;12(1):83. DOI: 10.3390/v12010083
42. Wang W, Tang J, Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol. 2020;92(4):441-447. DOI: 10.1002/jmv.25689
43. Malinnikova E.Yu. New coronaviral infection. Today’s look at the pandemic of the XXI century. Infektsionnye bolezni: novosti, mneniya, obuchenie [Infectious Diseases: News, Opinions, Training]. 2020;9(2):18-32. (In Russ.). DOI: 10.33029/2305-3496-2020-9-2-18-32
44. van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, et al. Aerosol and Surface Stability of SARSCoV-2 as Compared with SARS-CoV-1. N Engl J Med. 2020;382(16):1564-1567. DOI: 10.1056/NEJMc2004973
45. Lu CW, Liu XF, Jia ZF. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395(10224):e39. DOI: 10.1016/S0140-6736(20)30313-5
About the Authors
A. A. KononenkoRussian Federation
Anna A. Kononenko, Junior Researcher of the Laboratory of Sanitary Protection of the territory
Rostov-on-Don
A. K. Noskov
Russian Federation
Alexey K. Noskov, Cand. Sci. (Med), Director
Rostov-on-Don
S. Yu. Vodyanitskaya
Russian Federation
Svetlana Y. Vodyanitskaya, Can. Sci (Med), Chief specialist (infectious disease specialist) Department of Medical Care for Adults of the Department of Medical and Preventive Care Ministry of Health RO
Rostov-on-Don
O. A. Podoynitsyna
Russian Federation
Oksana A. Podoinitsyna, Cand. Sci (Bio), Researcher at the Laboratory of Cholera Microbiology
Rostov-on-Don
Review
For citations:
Kononenko A.A., Noskov A.K., Vodyanitskaya S.Yu., Podoynitsyna O.A. Human coronaviruses that can cause emergencies. Medical Herald of the South of Russia. 2021;12(1):14-23. https://doi.org/10.21886/2219-8075-2021-12-1-14-23







































